产品中心
联系我们
销售专用:
地址:北京市海淀区西小口路66号中关村东升科技园C-1楼三层

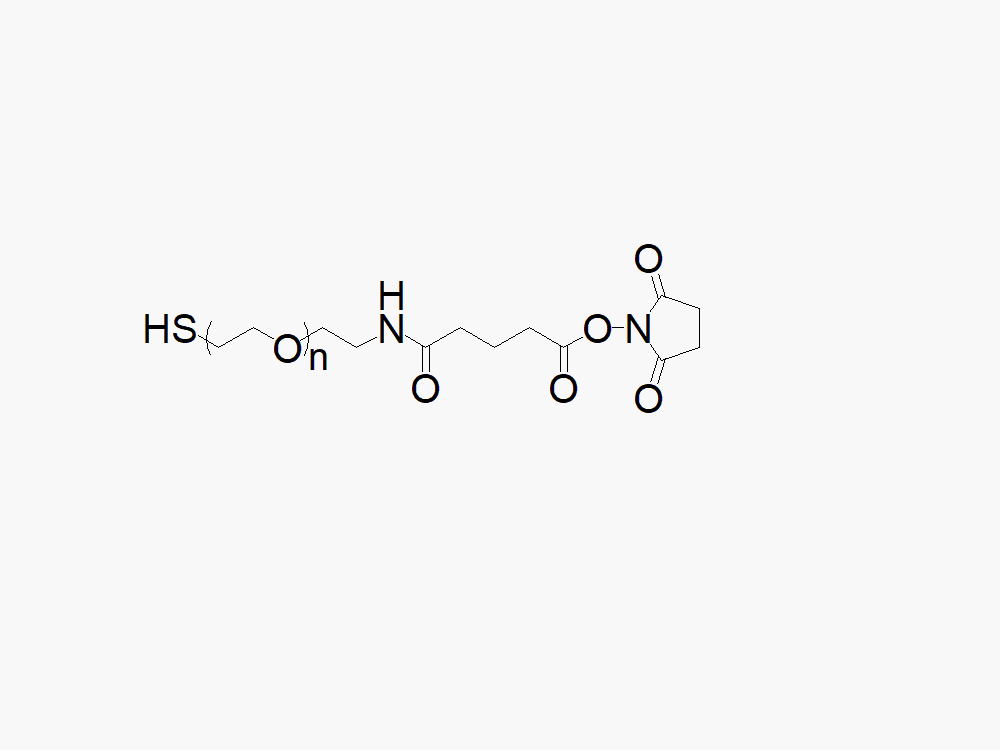
Biotin PEG Succinimidyl Carboxymethyl Ester
产品代号:
BIOTIN-PEG-SCM
产品纯度:
≥ 90%
包装规格:
1g, 10g, 100g等(特殊包装需收取分装费用)
分子量:
2000 Da,3500 Da, 5000 Da, 7500 Da等
关键词:
所属分类:
科研客户小批量一键采购地址(小于5克)
- 产品描述
- 参考文献
-
必赢网址bwi437科技提供高品质生物素聚乙二醇琥珀酰亚胺羧甲基酯,产品取代率>90%。
必赢网址bwi437科技生产的异双功能生物素PEG琥珀酰亚胺基羧甲基酯(SCM)产品通常用作两种不同化学物质的交联剂或间隔物。此异功能PEG衍生物中的PEG部分可提供水溶性、生物相容性及柔性。此产品专门应用于抗体偶联药物(ADC’s)的开发。
必赢网址bwi437科技提供BIOTIN-PEG-SCM分子量2000 Da,3500 Da, 5000 Da, 7500 Da的产品1克和10克包装。
必赢网址bwi437科技提供分装服务,需要收取分装费用,如果您需要分装为其他规格请与我们联系。
必赢网址bwi437科技同时提供其他分子量的BIOTIN-PEG-SCM衍生物产品,如你需要请与我司sales@jenkem.com联系。
必赢网址bwi437科技提供大批量生产产品及GMP级别产品,如需报价请与我们联系。
-
References:
1. Balzer, C.J., et al., Dip1 Co-opts Features of Branching Nucleation to Create Linear Actin Filaments that Activate WASP-Bound Arp2/3 Complex, Current Biology, 2018, 28(23), P. 3886-3891.
2. Li, M., Mapping Membrane Proteins on Cell Surface by AFM, Investigations of Cellular and Molecular Biophysical Properties by Atomic Force Microscopy, Nanorobotics, 2018, pp. 65-77.
3. Yu, Y., et al., A new NIR-triggered DOX and ICG co-delivery system for enhanced multidrug resistant cancer treatment through simultaneous chemo/photothermal/photodynamic therapy, Acta Biomaterialia, 2017.
4. Son, Y.J., et al., Electrospun Nanofibrous Sheets for Selective Cell Capturing in Continuous Flow in Microchannels, Biomacromolecules, 2016.
5. Zhang, W., et al., pH and near-infrared light dual-stimuli responsive drug delivery using DNA-conjugated gold nanorods for effective treatment of multidrug resistant cancer cells. Journal of Controlled Release, 2016, 232:9-19.
6. Li, M., et al., Rapid recognition and functional analysis of membrane proteins on human cancer cells using atomic force microscopy, Journal of Immunological Methods, 2016.
7. Chen, Y., et al., Force regulated conformational change of integrin αVβ3, Matrix Biology, 2016.
8. Oztug Durer, Z.A., et al., Metavinculin Tunes the Flexibility and the Architecture of Vinculin-Induced Bundles of Actin Filaments, Journal of Molecular Biology, 2015, 427:17, P. 2782-2798.
9. Rasson, A.S., et al., Filament Assembly by Spire: Key Residues and Concerted Actin Binding, Journal of Molecular Biology, 2015, 427 (4), p: 824-839.
10. Chen, Y., et al., Fluorescence Biomembrane Force Probe: Concurrent Quantitation of Receptor-ligand Kinetics and Binding-induced Intracellular Signaling on a Single Cell, J. Vis. Exp., 2015, 102, e52975.
11. Leiske, D.L., et al., Single-molecule enzymology based on the principle of the Millikan oil drop experiment, Analytical Biochemistry, 2014, 448, p: 30-37.
12. Petek, N.A. and R.D. Mullins, Chapter Two – Bacterial Actin-Like Proteins: Purification and Characterization of Self-Assembly Properties, in Methods in Enzymology, 2014, p. 19-34.
13. Zhao, G., et al., Functional PEG–PAMAM-Tetraphosphonate Capped NaLnF4 Nanoparticles and their Colloidal Stability in Phosphate Buffer. Langmuir, 2014, 30(23): p. 6980-6989.
14. Kisley, L., et al., Extending single molecule fluorescence observation time by amplitude-modulated excitation, Methods Appl. Fluoresc., 2013, 1 037001, 7pp.
15. Hansen, S.D, et al., Cytoplasmic actin: purification and single molecule assembly assays. InAdhesion Protein Protocols, 2013, pp. 145-170.
16. Scott, M.A., Ultra-rapid 2-D and 3-D laser microprinting of proteins. Diss. Massachusetts Institute of Technology, 2013.
17. Bor, B., et al., Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains, Mol. Biol. Cell, 2012, 23(19) 3801-3813.
18. Scott, M.A., Ultra-rapid laser protein micropatterning: screening for directed polarization of single neurons, Lab Chip, 2011.
19. Hansen, S.D., et al., VASP is a processive actin polymerase that requires monomeric actin for barbed end association, J. Cell Biol., 2010, 191(3) p: 571–584.
20. Andoy, N.M., et al., Single-Molecule Study of Metalloregulator CueR-DNA Interactions Using Engineered Holliday Junctions, Biophysical Journal, 97(3), 2009, p: 844-852.
21. Chen, X., et al., Using Aptamer-Conjugated Fluorescence Resonance Energy Transfer Nanoparticles for Multiplexed Cancer Cell Monitoring, Analytical Chemistry, 2009, 81(16) p: 7009–7014.
22. Ensign, A.E., Studies of Horse Heart Cytochrome c Folding, University of Rochester, 2009.
23. Ochs, C.J., et al., Low-Fouling, Biofunctionalized, and Biodegradable Click Capsules, Biomacromolecules, 2008, 9(12) p: 3389–3396.
24、Narvaez-Ortiz, H. Y., et al., Unconcerted conformational changes in Arp2/3 complex integrate multiple activating signals to assemble functional actin networks, Current Biology, 2022, 32(5).
25. Narvaez-Ortiz, HY, et al., Unconcerted conformational changes in Arp2/3 complex integrate multiple activating signals to assemble functional actin networks. Current Biology. 2022.
26. Rosado, AM, et al., Memory in Repetitive Protein–Protein Interaction Series—in Memory of the Late Professor Robert M. Nerem. bioRxiv. 2022.
产品询价